Proteomics Signaling Networks
Proteomics Signaling Networks
Our laboratory has developed quantitative proteomics approaches for systems biology analyses in green algae, plant and microbial systems. Due to their critical roles in regulation of cellular signaling, we have focused on approaches to explore proteome-wide post-translational modifications such as reversible oxidation, phosphorylation, and proteolysis.
Thiol-based Regulatory Switches:
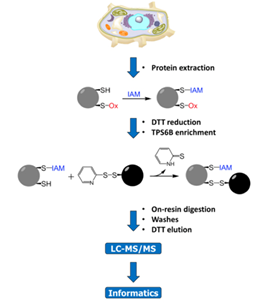
The influence of the redox environment as a regulatory factor in cellular processes has been present since the origin of life but is only beginning to be examined in detail. With a focus on the identification of protein targets of oxidation, the respective modification type, site specificity, quantification, and functional consequence, the field of redox biology remains highly complex and requires multiple approaches for systematic characterization. We routinely implement global and site-specific redox proteomics characterization methods by coupling an optimized reproducible disulfide-bond exchange resin enrichment (TPS6B) at the peptide- or protein-level with label-free quantitation via LC-MS/MS to elucidate relative redox changes in complex protein mixtures. We have coupled this with other PTM enrichments to facilitate the identification and elucidation of crosstalk between essential signaling pathway networks. Identification of redox proteins serves as a crucial element in understanding stress response in photosynthetic organisms, tardigrades and beyond.
Phosphoproteomics:
Protein phosphorylation, encompassing the addition or removal of phosphate moieties on the functional groups of amino acid side chains (typically Ser, Thr, Tyr), is ubiquitously found in eukaryotes and revealed to be critical for growth, adaptation and survival in photosynthetic organisms. Catalyzed by a class of enzymes known as kinases, these phosphorylation events serve as activators of protein function within the cell through induction of conformational change, and these kinases form networks that propagate phosphorylation signals through specific biological pathways as a mechanism for generating cellular responses from various stimuli.
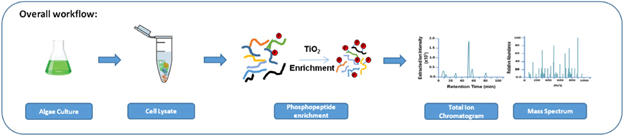
The study of these phosphorylation events in the cellular proteome, or phosphoproteomics, in photosynthetic organisms has the potential to not only inform on how life is sustained but also how an organism responds to their environment, with insights into broad ranging fields from agriculture, biofuels, and medicine. Our lab seeks to elucidate these phosphorylation events through complementary in vivo and in vitro platforms that utilize enrichment strategies specific to phosphorylation coupled with liquid chromatography and high mass accuracy, high resolution mass spectrometry (LC-MS) for broad, quantitative coverage of the phosphoproteome. Using enrichment strategies coupled with label-free quantitative proteomics, we have deeply interrogated the dynamic proteome and phosphoproteome of the model algae Chlamydomonas reinhardtii with >13 publications.
Proteolytic Pathways Involved in Stress Signaling Responses:
Plants are dynamic systems wherein external and internal signals are integrated to facilitate and sustain life. The intricate balance of protein translation and degradation, known as protein homeostasis, is key to maintaining proper cellular function. Despite its unequivocal importance, plant protein homeostasis and its intersection with stress signaling is relatively unexplored. Furthering our understanding of proteolysis in the plant stress response could impact the development of more adaptive and resistant crops.
Microbiome:
The role of the microbiome in human health and disease is well documented. Commensal bacteria produce complex metabolites to compete or protect against neighboring species; however, their roles in niche protection and host physiology, susceptibility to bioactive peptides introduced through diet, and connections between intra-species interactions are nascent.

Publications
A. L. Smythers et al., Chemobiosis reveals tardigrade tun formation is dependent on reversible cysteine oxidation. Plos One 19, e0295062 (2024).
P. W. Sadecki et al., Evolution of Polymyxin Resistance Regulates Colibactin Production in Escherichia coli. Acs Chem Biol 16, 1243-1254 (2021).
A. A. Iannetta et al., Profiling thimet oligopeptidase-mediated proteolysis in Arabidopsis thaliana. Plant J 106, 336-350 (2021).
A. A. Iannetta et al., IreK-Mediated, Cell Wall-Protective Phosphorylation in Enterococcus faecalis. J Proteome Res 20, 5131-5144 (2021).
A. L. Smythers, A. A. Iannetta, L. M. Hicks, Crosslinking mass spectrometry unveils novel interactions and structural distinctions in the model green alga Chlamydomonas reinhardtii. Mol Omics, (2021).
I. Couso et al., Inositol polyphosphates and target of rapamycin kinase signalling govern photosystem II protein phosphorylation and photosynthetic function under light stress in Chlamydomonas. New Phytol, (2021).

